The end of the year is a good time to recap and make predictions. 2020 has turned Graph ML into a celebrity of machine learning. For this post, I sought the opinion of prominent researchers in the field of graph ML and its applications trying to summarise the highlights of the past year and predict what is in store for 2021.
Beyond message passing
Will Hamilton, Assistant Professor at McGill University and CIFAR Chair at Mila, author of GraphSAGE.
“2020 saw the field of Graph ML come to terms with the fundamental limitations of the message-passing paradigm.
These limitations include the so-called “bottleneck” issue [1], problems with over-smoothing [2], and theoretical limits in terms of representational capacity [3,4]. Looking forward, I expect that in 2021 we will be searching for the next big paradigm for Graph ML. I am not sure what exactly the next generation of Graph ML algorithms will look like, but I am confident that progress will require breaking away from the message-passing schemes that dominated the field in 2020 and before.
I am also hopeful that 2021 will also see Graph ML move into more impactful and challenging application domains. Too much recent research focuses on simple, homophilous node-classification tasks. I also hope to see methodological advancements towards tasks that require more complex algorithmic reasoning, such as tasks involving knowledge graphs, reinforcement learning, and combinatorial optimisation.”
Algorithmic reasoning
Petar Veličković, Senior Researcher at DeepMind, author of Graph Attention Networks.
“2020 has definitively and irreversibly turned graph representation learning into a first-class citizen in ML.”
The great advances made this year are far too many to enumerate briefly, but I am personally most excited about neural algorithmic reasoning. Neural networks are traditionally very powerful in the interpolation regime, but are known to be terrible extrapolators — and hence inadequate reasoners; as one of the main traits of reasoning is being able to function out-of-distribution. Reasoning tasks are likely to be ideal for further development of GNNs, not only because they are known to align very well with such tasks [5], but also because many real-world graph tasks exhibit homophily, meaning that the most impactful and scalable approaches typically will be much simpler forms of GNNs [6,7].
Building on the historical successes of previous neural executors such as the Neural Turing Machine [8] and the Differentiable Neural Computer [9], and reinforced by the now-omnipresent graph machine learning toolbox, several works published in 2020 explored the theoretical limits of neural executors [5,10,11], derived novel and stronger reasoning architectures based on GNNs [12–15], and enabled perfect strong generalisation on neural reasoning tasks [16]. While such architectures could naturally translate into wins for combinatorial optimisation [17] in 2021, I am personally most thrilled about how pre-trained algorithmic executors can allow us to apply classical algorithms to inputs that are too raw or otherwise unsuitable for the algorithm. As one example, our XLVIN agent [18] used exactly these concepts to allow a GNN to execute value iteration style algorithms within the reinforcement learning pipeline, even though the specifics of the underlying MDP were not known. I believe 2021 will be ripe with GNN applications to reinforcement learning in general.”
Relational structure discovery
Thomas Kipf, Research Scientist at Google Brain, author of Graph Convolutional Networks.
“One particularly noteworthy trend in the Graph ML community since the recent widespread adoption of GNN-based models is the separation of computation structure from the data structure.
In a recent ICML workshop talk, I termed this trend relational structure discovery. Typically, we design graph neural networks to pass messages on a fixed (or temporally evolving) structure provided by the dataset, i.e. the nodes and edges of the dataset are taken as the gold standard for the computation structure or message passing structure of our model.
In 2020, we have seen rising interest in models that are able to adapt the computation structure, i.e., which components they use as nodes and over which pairs of nodes they perform message passing, on the fly — while going beyond simple attention-based models. Influential examples in 2020 include Amortised Causal Discovery [19–20], which makes use of Neural Relational Inference to infer (and reason with) causal graphs from time-series data, GNNs with learnable pointer [21,15] and relation mechanisms [22–23], learning mesh-based physical simulators with adaptive computation graphs [24], and models that learn to infer abstract nodes over which to perform computations [25–26]. This development has widespread implications, as it allows us to effectively utilise symmetries (e.g. node permutation equivariance) and inductive biases (e.g. modeling of pairwise interaction functions) afforded by GNN architectures in other domains, such as text or video processing.
Going forward, I expect that we will see many developments in how one can learn the optimal computational graph structure (both in terms of nodes and relations) given some data and tasks without relying on explicit supervision. Inspection of such learned structures will likely be valuable in deriving better explanations and interpretations of the computations that learned models perform to solve a task, and will likely allow us to draw further analogies to causal reasoning.”
Expressive power
Haggai Maron, Research Scientist at Nvidia, author of provably expressive high-dimensional graph neural networks.
“The expressive power of graph neural networks was one of the central topics in Graph ML in 2020.
There were many excellent papers discussing the expressive power of various GNN architectures [27] and showing fundamental expressivity limits of GNNs when their depth and width is restricted [28], describing what kind of structures can be detected and counted using GNNs [29], showing that using a fixed number of GNNs does not make sense for many graph tasks and suggesting an iterative GNN that learns to terminate the message passing process adaptively [14].
In 2021, I would be happy to see advancements in principled approaches for generative models for graphs, connections between graph matching with GNNs and the expressive power of GNNs, learning graphs of structured data like images and audio, and developing stronger connections between the GNN community and the computer vision community working on scene graphs.”
Scalability
Matthias Fey, PhD student at TU Dortmund, developer of PyTorch Geometric and Open Graph Benchmark.
“One of the most trending topics in Graph ML research in 2020 was tackling the scalability issues of GNNs.
Several approaches relied on simplifying the underlying computation by decoupling prediction from propagation. We have seen numerous papers that simply combine a non-trainable propagation scheme with a graph-agnostic module, either as a pre- [30,7] or post-processing [6] step. This leads to superb runtime and, remarkably, mostly on par performance on homophily graphs. With access to increasingly bigger datasets, I am eager to see how to advance from here and how to make use of trainable and expressive propagation in a scalable fashion.”
Dynamic graphs
Emanuele Rossi, ML Researcher at Twitter and PhD student at Imperial College London, author of Temporal Graph Networks.
“Many interesting Graph ML applications are inherently dynamic, where both the graph topology and the attributes evolve over time.
This is the case in social networks, financial transaction networks, or user-item interaction networks. Until recently, the vast majority of research on Graph ML has focused on static graphs. The few works attempting to deal with dynamic graphs mainly considered discrete-time dynamic graphs, a series of graph snapshots at regular intervals. In 2020, we saw an emerging set of works [31–34] on a more general category of continuous-time dynamic graphs, that can be thought of as an asynchronous stream of timed events. Moreover, the first interesting successful applications of models for dynamic graphs are also starting to emerge: we saw fake account detection [35], fraud detection [36], and controlling the spreading of an epidemic [37].
I think that we are only scratching the surface of this exciting direction and many interesting questions remain unanswered. Among important open problems are scalability, better theoretical understanding of dynamic models, and combining spatial and temporal diffusion of information in a single framework. We also need more reliable and challenging benchmarks to make sure progress can be better evaluated and tracked. Finally, I hope to see more successful applications of dynamic graph neural architectures, especially in the industry.”
New hardware
Mark Saroufim, ML Engineer at Graphcore.
“I cannot think of a single customer I have worked with who has not either deployed a Graph Neural Network in production or is planning to.
Part of this trend is that the natural graph structure in applications such as NLP, protein design, or molecule property prediction has been traditionally ignored, and instead the data was treated as sequences amenable for existing and well-established ML models such as Transformers. We know, however, that Transformers are nothing but GNNs where attention is used as the neighbourhood aggregation function. In computing, the phenomenon when certain algorithms win not because they are ideally suited to solve a certain problem, but because they run well on existing hardware is called Hardware Lottery [38] — and this is the case with Transformers running on GPUs.
At Graphcore, we have built a new MIMD architecture with 1472 cores that can run a total of 8832 programs in parallel, which we call the Intelligence Processing Unit (IPU). This architecture is ideally suited for accelerating GNNs. Our Poplar software stack takes advantage of sparsity to allocate different nodes of a computational graph to different cores. For models that can fit into the IPU’s 900MB on-chip memory, our architecture offers substantial improvement of the throughput over GPUs; otherwise, with just a few lines of code, it is possible to distribute the model over thousands of IPUs.
I am excited to see our customers building a large body of research taking advantage of our architecture, including applications such as bundle adjustment for SLAM, training deep networks using local updates, or speeding up a variety of problems in particle physics. I hope to see more researchers taking advantage of our advanced ML hardware in 2021.”
Applications in the industry, physics, medicine, and beyond

Sergey Ivanov, Research Scientist at Criteo, editor of the Graph Machine Learning newsletter.
“It was an astounding year for Graph ML research. All major ML conferences had about 10–20% of all papers dedicated to this field and at this scale, everyone can find an interesting graph topic to their taste.
The Google Graph Mining team was prominently present at NeurIPS. Looking at the 312-page presentation, one can say that Google has advanced in utilising graphs in production more than anyone else. The applications they address using Graph ML include modeling COVID-19 with spatio-temporal GNNs, fraud detection, privacy preservation, and more. Furthermore, DeepMind rolled out GNNs in production for travel time predictions globally in Google Maps. An interesting detail of their method is the integration of an RL model to select similar sampled subgraphs into a single batch for training parameters of GNNs. This and advanced hyperparameter tuning brought up to +50% improvement in the accuracy of real-time time-of-arrival estimation.
Another notable application of GNNs was done at Magic Leap, which specialises in 3D computer-generated graphics. Their SuperGlue architecture [39] applies GNNs to feature matching in images — an important subject for 3D reconstruction, place recognition, localisation, and mapping. This end-to-end feature representation paired with optimal transport optimisation triumphed on real-time indoor and outdoor pose estimation. These results just scratch the surface of what has been achieved in 2020.
Next year, I believe we will see further use of Graph ML developments in industrial settings. This would include production pipelines and frameworks, new open-source graph datasets, and deployment of GNNs at scale for e-commerce, engineering design, and the pharmaceutical industry.”
Kyle Cranmer, Professor of Physics at NYU, one of the discoverers of the Higgs boson.
“It has been amazing to see how in the last two years Graph ML has become very popular in the field of physics.
Early work with deep learning in particle physics often forced the data into an image representation to work with CNNs, which was not natural as our data are not natively grid-like and the image representation is very sparse. Graphs are a much more natural representation of our data [40,41]. Researchers on the Large Hadron Collider are now working to integrate Graph ML into the real-time data processing systems that process billions of collisions per second. There is an effort to achieve this by deploying inference servers to integrate Graph ML with the real-time data acquisition systems [42] and efforts to implement these algorithms on FPGAs and other special hardware [43].
Another highlight from Graph ML in 2020 is the demonstration that its inductive bias can pair with symbolic approaches. For example, we used a GNN to learn how to predict various dynamical systems, and then we ran symbolic regression on the messages being sent along the edges [44]. Not only were we able to recover the ground-truth force laws for those dynamical systems, but we were also able to extract equations in situations where we don’t have ground truth. Amazingly, the symbolic equations that were extracted could then be re-introduced into the GNN, replacing the original learned components, and we obtained even better generalisation to out of distribution data.”
Anees Kazi, PhD student at TUM, author of multiple papers on Graph ML in medical imaging.
“In the medical domain, Graph ML transformed the way of analyzing multimodal data in a way that closely resembles how experts look at the patient’s condition from all the available dimensions in clinical routines.
There has recently been a huge spike in the research related to Graph ML in medical imaging and healthcare applications [45], including brain segmentation [46], brain structure analysis using MRI/fMRI data targeted towards disease prediction [47], and drug effect analysis [48].
Among topics in Graph ML, several stood out in the medical domain in 2020. First, latent graph learning [22,49,50], as empirically defining a graph for the given data was till then a bottleneck for optimal outcomes, now has been solved by methods which learn the latent graph structure automatically. Second, data imputation [51], as missing data is one standing problem in a lot of datasets in the medical domain, graph-based methods have helped in the imputation of data depending on relations coming from graph neighbourhood. Third, the interpretability for Graph ML models [52], since it is important for clinical and technical experts to focus on reasoning the outcomes of Graph ML models for their reliable incorporation into a CADx system. Another important highlight of 2020 in the medical domain was of course the coronavirus pandemic, and Graph ML methods were used for detection of Covid-19 [53].
In 2021, Graph ML could be used to further the interpretability of ML models for better decision making. Secondly, it has been observed that Graph ML methods are still sensitive to the graph structure, hence robustness to graph perturbations and adversarial attacks is an important topic. Finally, it would be interesting to see the integration of self-supervised learning with Graph ML applied to the medical domain.”
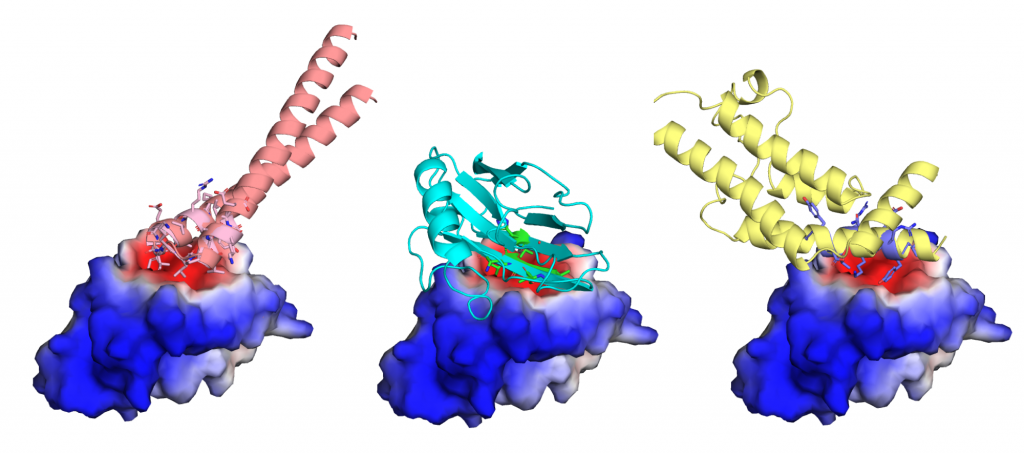
Bruno Correia, Assistant Professor at EPFL, head of the Protein Design and Immunoengineering Laboratory, one of the developers of MaSIF.
“In 2020, exciting progress has been made in protein structure prediction, a key problem in bioinformatics. Yet, ultimately the chemical and geometric pattern displayed at the surface of these molecules are critical for protein function.
Surface-based representations of molecules have been used for decades but they pose challenges for machine learning methods. Approaches from the realm of Geometric Deep Learning have brought impressive capabilities to the field of protein modeling given their ability to deal with irregular data, which are particularly well-suited for protein representations. In MaSIF [1], we used geometric deep learning on mesh-based molecular surface representations to learn patterns that allow us to predict interactions of proteins with other molecules (proteins and metabolites) and speed up docking calculations by several orders of magnitude. This, in turn, could facilitate a much larger scale of prediction of protein-protein interaction networks.
In a further development of the MaSIF framework [2], we managed to generate our surface and chemical feature on the fly avoiding all precomputation stages. I anticipate that such advances will be transformative for protein and small molecule design, and in the long term could help faster development of biological drugs.”
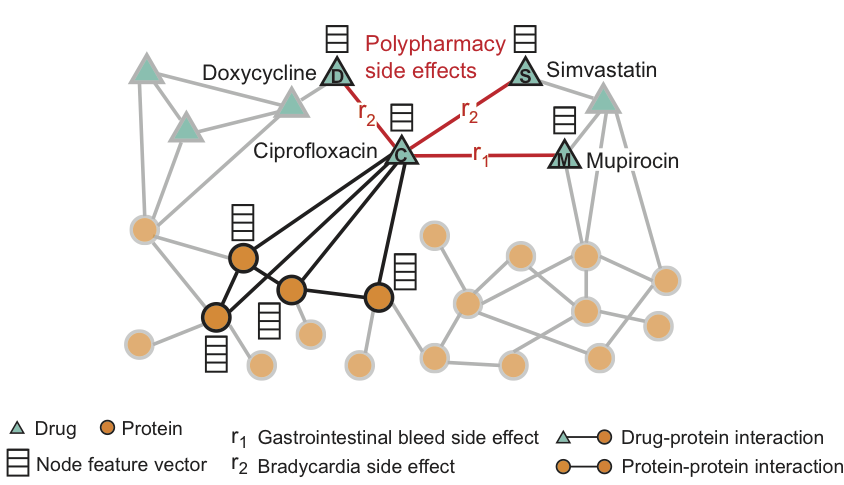
Marinka Zitnik, Assistant Professor of Biomedical Informatics at Harvard Medical School, author of Decagon.
“It was exciting to see how Graph ML entered the fields of life sciences in 2020.
We have seen how graph neural networks not only outperform earlier methods on carefully designed benchmark datasets but can open up avenues for developing new medicines to help people and understanding nature at the fundamental level. Highlights include advances in single-cell biology [56], protein and structural biology [54,57], and drug discovery[58] and repositioning [59].
For centuries, the scientific method — the fundamental practice of science that scientists use to systematically and logically explain the natural world — has remained largely the same. I hope that in 2021, we will make substantial progress on using Graph ML to change that. To do that, I think we need to design methods that can optimize and manipulate networked systems and predict their behavior, such as how genomics — Nature’s experiments on people — influences human traits in the context of disease. Such methods need to work with perturbational and interventional data (not only ingest observational measurements of our world). Also, I hope we will develop more methods for learning actionable representations that readily lend themselves to actionable hypotheses in science. Such methods can enable decision making in high-stakes settings (e.g., chemistry tests, particle physics, human clinical trials) where we need precise, robust predictions that can be interpreted meaningfully.”
[1] U. Alon and E. Yahav, On the bottleneck of graph neural networks and its practical implications (2020) arXiv:2006.05205.
[2] Q. Li, Z. Han, X.-M. Wu, Deeper insights into graph convolutional networks for semi-supervised learning (2019) Proc. AAAI.
[3] K. Xu et al. How powerful are graph neural networks? (2019) Proc. ICLR.
[4] C. Morris et al. Weisfeiler and Leman go neural: Higher-order graph neural networks (2019) Proc. AAAI.
[5] K. Xu et al. What can neural networks reason about? (2019) arXiv:1905.13211.
[6] Q. Huang et al. Combining label propagation and simple models out-performs graph neural networks (2020) arXiv:2010.13993.
[7] F. Frasca et al. SIGN: Scalable Inception Graph Neural Networks (2020) arXiv:2004.11198.
[8] A. Graves, G. Wayne, and I. Danihelka, Neural Turing Machines (2014) arXiv:1410.5401.
[9] A. Graves et al. Hybrid computing using a neural network with dynamic external memory (2016). Nature 538:471–476.
[10] G. Yehuda, M. Gabel, and A. Schuster. It’s not what machines can learn, it’s what we cannot teach (2020) arXiv:2002.09398.
[11] K. Xu et al. How neural networks extrapolate: From feedforward to graph neural networks (2020) arXiv:2009.11848.
[12] P. Veličković et al., Neural execution of graph algorithms (2019) arXiv:1910.10593.
[13] O. Richter and R. Wattenhofer, Normalized attention without probability cage (2020) arXiv:2005.09561.
[14] H. Tang et al., Towards scale-invariant graph-related problem solving by iterative homogeneous graph neural networks (2020) arXiv:2010.13547.
[15] P. Veličković et al. Pointer Graph Networks (2020) Proc. NeurIPS.
[16] Y. Yan et al. Neural execution engines: Learning to execute subroutines (2020) Proc. ICLR.
[17] C. K. Joshi et al. Learning TSP requires rethinking generalization (2020) arXiv:2006.07054.
[18] A. Deac et al. XLVIN: eXecuted Latent Value Iteration Nets (2020) arXiv:2010.13146.
[19] S. Löwe et al., Amortized Causal Discovery: Learning to infer causal graphs from time-series data (2020) arXiv:2006.10833.
[20] Y. Li et al., Causal discovery in physical systems from videos (2020) Proc. NeurIPS.
[21] D. Bieber et al., Learning to execute programs with instruction pointer attention graph neural networks (2020) Proc. NeurIPS.
[22] A. Kazi et al., Differentiable Graph Module (DGM) for graph convolutional networks (2020) arXiv:2002.04999
[23] D. D. Johnson, H. Larochelle, and D. Tarlow., Learning graph structure with a finite-state automaton layer (2020). arXiv:2007.04929.
[24] T. Pfaff et al., Learning mesh-based simulation with graph networks (2020) arXiv:2010.03409.
[25] T. Kipf et al., Contrastive learning of structured world models (2020) Proc. ICLR
[26] F. Locatello et al., Object-centric learning with slot attention (2020) Proc. NeurIPS.
[27] W. Azizian and M. Lelarge, Characterizing the expressive power of invariant and equivariant graph neural networks (2020) arXiv:2006.15646.
[28] A. Loukas, What graph neural networks cannot learn: depth vs width (2020) Proc. ICLR.
[29] Z. Chen et al., Can graph neural networks count substructures? (2020) Proc. NeurIPS.
[30] A. Bojchevski et al., Scaling graph neural networks with approximate PageRank (2020) Proc. KDD.
[31] E. Rossi et al., Temporal Graph Networks for deep learning on dynamic graphs (2020) arXiv:2006.10637.
[32] S. Kumar, X. Zhang, and J. Leskovec, Predicting dynamic embedding trajectory in temporal interaction networks (2019) Proc. KDD.
[33] R. Trivedi et al., DyRep: Learning representations over dynamic graphs (2019) Proc. ICLR.
[34] D. Xu et al., Inductive representation learning on temporal graphs (2019) Proc. ICLR.
[35] M. Noorshams, S. Verma, and A. Hofleitner, TIES: Temporal Interaction Embeddings for enhancing social media integrity at Facebook (2020) arXiv:2002.07917.
[36] X. Wang et al., APAN: Asynchronous Propagation Attention Network for real-time temporal graph embedding (2020) arXiv:2011.11545.
[37] E. A. Meirom et al., How to stop epidemics: Controlling graph dynamics with reinforcement learning and graph neural networks (2020) arXiv:2010.05313.
[38] S. Hooker, Hardware lottery (2020), arXiv:2009.06489.
[39] P. E. Sarlin et al., SuperGlue: Learning feature matching with graph neural networks (2020). Proc. CVPR.
[40] S. Ruhk et al., Learning representations of irregular particle-detector geometry with distance-weighted graph networks (2019) arXiv:1902.07987.
[41] J. Shlomi, P. Battaglia, J.-R. Vlimant, Graph Neural Networks in particle physics (2020) arXiv:2007.13681.
[42] J. Krupa et al., GPU coprocessors as a service for deep learning inference in high energy physics (2020) arXiv:2007.10359.
[43] A. Heintz et al., Accelerated charged particle tracking with graph neural networks on FPGAs (2020) arXiv:2012.01563.
[44] M. Cranmer et al., Discovering symbolic models from deep learning with inductive biases (2020) arXiv:2006.11287. Miles Cranmer is unrelated to Kyle Cranmer, though both are co-authors of the paper. See also the video presentation of the paper.
[45] Q. Cai et al., A survey on multimodal data-driven smart healthcare systems: Approaches and applications (2020) IEEE Access 7:133583–133599
[46] K. Gopinath, C. Desrosiers, and H. Lombaert, Graph domain adaptation for alignment-invariant brain surface segmentation (2020)arXiv:2004.00074
[47] J. Liu et al., Identification of early mild cognitive impairment using multi-modal data and graph convolutional networks (2020) BMC Bioinformatics 21(6):1–12
[48] H. E. Manoochehri and M. Nourani, Drug-target interaction prediction using semi-bipartite graph model and deep learning (2020). BMC Bioinformatics 21(4):1–16
[49] Y. Huang and A. C. Chung, Edge-variational graph convolutional networks for uncertainty-aware disease prediction (2020) Proc. MICCAI
[50] L. Cosmo et al., Latent-graph learning for disease prediction (2020) Proc. MICCAI
[51] G. Vivar et al., Simultaneous imputation and disease classification in incomplete medical datasets using Multigraph Geometric Matrix Completion (2020) arXiv:2005.06935.
[52] X. Li and J. Duncan, BrainGNN: Interpretable brain graph neural network for fMRI analysis (2020) bioRxiv:2020.05.16.100057
[53] X. Yu et al., ResGNet-C: A graph convolutional neural network for detection of COVID-19 (2020) Neurocomputing.
[54] P. Gainza et al., Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning (2020) Nature Methods 17(2):184–192.
[55] F. Sverrisson et al., Fast end-to-end learning on protein surfaces (2020) bioRxiv:2020.12.28.424589.
[56] A. Klimovskaia et al., Poincaré maps for analyzing complex hierarchies in single-cell data (2020) Nature Communications 11.
[57] J. Jumper et al., High accuracy protein structure prediction using deep learning (2020) a.k.a. AlphaFold 2.0 (paper not yet available).
[58] J. M. Stokes et al., A deep learning approach to antibiotic discovery (2020) Cell 180(4):688–702.
[59] D. Morselli Gysi et al., Network medicine framework for identifying drug repurposing opportunities for COVID-19 (2020) arXiv:2004.07229.
I am grateful to Bruno Correia, Kyle Cranmer, Matthias Fey, Will Hamilton, Sergey Ivanov, Anees Kazi, Thomas Kipf, Haggai Maron, Emanuele Rossi, Mark Saroufim, Petar Veličković, and Marinka Zitnik for their inspiring comments and predictions. This is my first experiment with a new format of “scientific journalism” and I appreciate suggestions for improvement. Needless to say, all the credit goes to the aforementioned people, whereas any criticism should be my sole responsibility. A Chinese translation of this post is available courtesy of Zhiqiang Zhong.